UNC School of Medicine scientists led by Scott Magness, PhD, sequenced the genes expressed in individual single cells from human digestive tracts to discover new cell-type characteristics and gain insights into important cell functions such as nutrient absorption and immune defense.
CHAPEL HILL, NC – If you get nervous, you might feel it in your gut. If you eat chili, your gut might revolt, but your friend can eat anything and feel great. You can pop ibuprofen like candy with no ill effects, but your friend’s belly might bleed and she might get no pain relief. Why is this? The quick answer is because we’re all different. The next questions are how different exactly, and what do these differences mean for health and disease? Answering these is much more difficult, but the UNC School of Medicine lab of Scott Magness, PhD, is revealing some interesting scientific answers.
For the first time, the Magness lab used entire human GI tracts from three organ donors to show how cell types differ across all regions of the intestines, to shed light on cellular functions, and to show gene expression differences between these cells and between individuals.
This work, published in Cellular and Molecular Gastroenterology and Hepatology, opens the door to exploring the many facets of gut health in a much more precise manner at greater resolution than ever before.
“Our lab showed it’s possible to learn about each cell type’s function in important processes, such as nutrient absorption, protection from parasites, and the production of mucus and hormones that regulate eating behavior and gut motility,” said Magness, associate professor in the Joint UNC-NC State Department of Biomedical Engineering and senior author of the paper. “We also learned how the gut lining might interact with the environment through receptors and sensors, and how drugs could interact with different cell types.”
The Sensitive Gut
Think of a typical pharmaceutical commercial voiceover when the voice actor pleasantly recites possible side effects, such as diarrhea, vomiting, intestinal bleeding, and other unpleasant collateral damage. Well, the Magness lab is attempting to understand why those side effects happen, down to the level of individual cells, their functions, their locations, and their genes.

For this research, the Magness lab focused on the epithelium: the single-cell thick layer separating the inside of the intestines and colon from everything else. Like other cell populations and the microbiota, the epithelium is incredibly important to human health, and for years scientists have been exploring it. But until now, researchers could only take tiny biopsies the size of grains of rice from a few parts of the digestive tract, usually from the colon or limited regions of the small intestine.
“Such exploration would be like looking at the United States from space but only investigating what’s going on in Massachusetts, Oklahoma, and California,” Magness said. “To really learn about the country, we’d want to see everything.”
Magness leaned on co-first authors, postdoctoral fellow Joseph Burclaff, PhD, and graduate student, Jarrett Bliton, both trainees in the Magness lab.

“Not only do we want to identify where the cells are located, but we want to know exactly which cell types do what, and why,” Burclaff said. “So, staying with the map analogy, we don’t want to just say, ‘oh, there’s North Carolina’. We want to know where to get the best barbecue. We want a ground level view to know as much as possible.”
In the past, researchers would mash up those rice-sized biopsies to identify all epithelial cell types and learn some general features of these cells. Magness’s approach was to sample thousands of individual cells from every part of the lower digestive tract (small intestine and colon) to create an atlas and then study the potential roles of these cells through the genes that each cell expresses. Knowing all of this would deepen scientific knowledge about the gut epithelium and hopefully encourage other scientists to explore each cell’s function in biology, in disease, and in the unfortunate scenario of pharmaceutical side effects.
To do such a deep individual cell dive, Magness needed two things: better technology and the entire digestive tracts of humans.
The Biology of Data
UNC-Chapel Hill acquired state-of-the-art RNA sequencing technology several years ago for the creation of the Advanced Analytics Core Facility through the UNC Center for Gastrointestinal Disease and Biology, which developed the scientific and intellectual heft – research faculty, staff, postdocs, and students – to use state-of-the-art equipment.
The Magness Group acquired human digestive tracts through a research agreement with organ donor services at HonorBridge. When intestines are harvested for transplant and if they are not claimed by higher-priority groups, HonorBridge staff coordinates with the Magness Group to donate the transplant-grade organs for research.
Six to eight hours after harvest, the Magness lab receives intact intestinal tracts, each about 15 to 30 feet long. They remove the epithelial layer, which is one long connected piece of tissue despite being only one cell thick. Then the researchers use enzymes to break down the epithelium into individual cells. For this study, they repeated this for organs from three separate donors.
Using sequencing technology to characterize gene expression, the Magness group first extracts RNA from each cell while keeping each cell separate, and then they run single-cell sequencing, which takes a snapshot of which genes each intestinal cell is expressing and how much.
“The picture we get from each cell is a mosaic of all the different types of genes the cells make and this complement of genes creates a ‘signature’ to tell us what kind of cell it is and potentially what it is doing,” Magness said. “Is it a stem cell or a mucous cell or a hormone-producing cell or an immune-signaling cell?
Burclaff added, “We were able to see the differences in cell types throughout the entire digestive tracts, and we can see different gene expression levels in the same cell types from three different people. We can see the different sets of genes turned on or off in individual cells. This is how, for instance, we might begin to understand why some people form toxicity to certain foods or drugs and some people don’t.”
A major problem with this kind of research is the sheer amount of data produced. The single cell sequencing picks up about 11,000 ‘reads’, or individual samples of gene products in just one cell, and in many thousands of individual cells, each with different combinations of the 20,000-plus human genes that are turned on or off. This creates almost 140,000,000 data points for all the 12,590 cells in the study that have to be put into a “visual” format so that scientists can make sense of the vast amount of information.
“The human brain can only comprehend two dimensions, three is challenging,” Magness said. “Add time, and it’s even trickier to comprehend what a single cell is up to. The amount of data our experiments produced was basically millions of dimensions all at once.”
Bliton devised computational techniques to filter the data to produce a manageable data set that included cell populations from all portions of the GI tract. Then, based on what Magness and other researchers had already learned of each cell type, Bliton could computationally identify each cell type from each region. He then plotted these data in a manner that humans can understand and interpret.
Reining in the immense data allowed the scientists to learn a lot about each cell type. Consider the tuft cell, discovered 40 years ago and so named

because they look as if they have tufts of hair on their surface. Turns out these tuft cells express similar genes as those on taste buds on the tongue. Other researchers discovered that these tuft cells sensed worm infections and sent signals to the immune system to begin waging war. The Magness lab showed that tuft cells exhibit a set of genes thought to be important for sensing and “tasting” other kinds of intestinal content so it can signal the immune system if need be. This would represent a much broader function than sensing if there’s a parasite in your gut or not.
“Not only did we describe every single cell type and every single gene they express individually, but we also looked at potential functions,” Burclaff said. “If you look at intestinal mucus, which is a complex mixture that protects the cells, we show which cells express various mucin proteins, how much, and in which regions of the digestive tract. We looked at where specific enzymes that digest food are expressed. We looked at cells with anti-inflammatory gene expression and synapse genes where the gut is probably connected to nerves so it can talk to the rest of the body. We looked at aquaporins, proteins involved in transferring water through the intestinal membrane.”
What the Magness group found was a whole new level of variation in potential functions that had not previously been appreciated through mashing up biopsy samples.
The researchers explored all epithelial receptors – the cell surface proteins used to communicate with other cells and molecules and with the environment of the gut. Magness and colleagues could see which receptors were expressed the most and in which cell types, painting a new picture of how cells might interact with gut contents such as nutrients, microbes, toxins, and drugs.
“As far as we know, we’re the first to do this kind of analysis across the length of the human gut from three full donors,” Bliton said. “We can look at each cell type and predict which pharmaceuticals might affect which cell type individually.”
For instance, there’s a class of drugs to treat inflammatory bowel disease; they’re designed to hit specific targets, certain immune cells that trigger inflammation. But the Magness lab learned that some epithelial cells express the same genes as those in the immune cells that are intended to be the target. This finding indicates there could be “off-target” effects in epithelial cells that are not intended and could lead to side-effects.
“This was not known,” Burclaff said. “Lots of drugs have bad GI side effects. And it could be because the drugs are affecting individual cells along the entire length of the GI tract. We show where these receptors are most expressed and in which cell types.”
This kind of knowledge is just one outcome from the Magness lab’s initial study.
“We want the scientific, medical, and pharmaceutical community to use what we’ve found,” Magness said. “We adopted an analytic approach to methodically address each cell type, produce easy-to-read and accessible spreadsheets for most scientists, and show several examples of what we can be discovered with this kind of high resolution, precision approach.”
Funding for this research came from the National Institutes of Health, the Katherine E. Bullard Charitable Trust, the Crohn’s and Colitis Foundation, and the University Cancer Research Fund at UNC-Chapel Hill.
Aside from the aforementioned researchers, other authors are Keith Breau, Meryem Ok, Ismael Gomez-Martinez, Jolene Ranek, Aadra Bhatt, Jeremy Purvis, and John Woosley, all at UNC-Chapel Hill.
UNC School of Medicine contact: Mark Derewicz
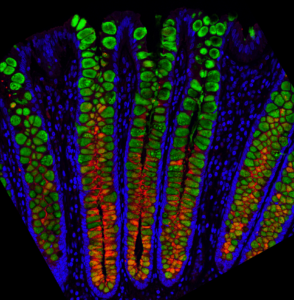
Image 1: The Magness Group fluorescently stained mucin proteins which contribute to the mucus layer that protects the colon. Here, thin sections of fixed human colon are stained for two different mucin proteins: MUC2 in green and MUC5B in red, with all nuclei in blue. This shows that the mucus content likely differs between the bottoms and the tops of the colon crypts and confirmed their results from the single cell gene sequencing.
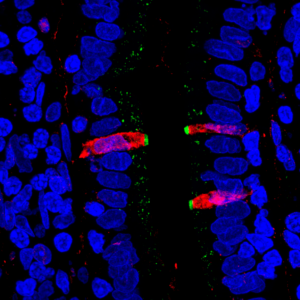
Image 2: The Magness group fluorescently stained proteins in thin section of human small intestine. They found that CFTR, a gene involved in cystic fibrosis (stained green), and FKBP1A, a target of pharmaceuticals intended to affect immune cells (stained red), are both present in a cell type within the human intestinal lining, confirming their results from the single cell gene sequencing.